



Porcine Circovirus Type 2: Genotypes and protection, where do we stand?
Porcine Circovirus type 2 (PCV2) has been an unusual virus ever since its emergence in the productive setting back in the 1990s.
Since its discovery, this virus has been observed to be involved in a wide variety of clinical presentations, such as reproductive and respiratory conditions and the dermatitis and nephropathy syndrome, apart from the post-weaning multisystemic wasting disease.
Despite this, the most common form of the disease is considered to be the subclinical (Segalés et al. 2012). The roles of PCV2 in these diseases, as well as its pathogenic mechanisms, are not entirely clear; hence, it is not so easy to interpret what the detection of the virus in animals or in tissue samples means or implies.
One of the most salient characteristics of this virus is its mutation rate, considered to be one of the highest for DNAss viruses (Firth, 2009). This finding helped to change the paradigm on the evolution of DNA viruses. In the specific case of PCV2, mutations have made the appearance of different genotypes possible. Another of the known factors is recombination, which contributes to virus’ diversification, this being a characteristic that is particularly important in the appearance of new types of virus.
In the case of PCV2, a number of cases of recombination have been described, although the reliability thereof is questioned by the scientific community (Franzo et al. 2018).
Accordingly, the contribution of recombination to the generation of new genotypes of PCV2 remains unclear.
The evolution of the virus has followed a constant pathway throughout its history. Its propensity to accumulate mutations has helped to generate a high degree of variability in its genome. Over time, different genotypes have appeared, although not all of them have persisted or spread among the porcine population. At present, the most accepted classification distinguishes between eight genotypes, ranging from PCV2a to PCV2h (Franzo, 2018).
Despite this, PCV2a, PCV2b and PCV2d are the most widespread genotypes, accounting for 96.31% of the sequenced PCV2 genomes in domestic pigs (Franzo, 2018).
The prevalence (and relevance) of these genotypes has changed constantly over time. PCV2a was the first genotype to appear, in 1996, and it dominated the
epidemiological setting until the change to PCV2b in 2003 (Dupont et al. 2007), which in turn was followed by the change to PCV2d in the same decade (Guo et al. 2010). Despite PCV2 having a global distribution, there are geographical differences in the distribution of its genotypes (Figure 1).
Of the 8 existing genotypes, only PCV2 a, b and d have a global distribution (Franzo et al. 2018); of these three, PCV2b is the one most isolated in Europe and the Americas. In turn, PCV2d was particularly relevant in Asia, where its isolation was greater than that of PCV2b.
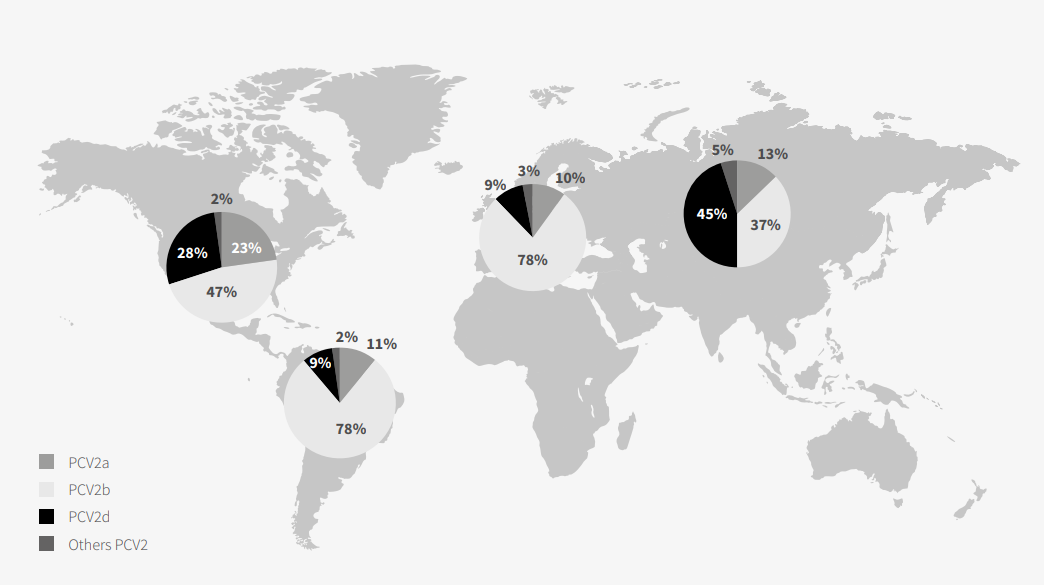
As has already been mentioned, PCV2a is currently the one with the lowest prevalence in the aforesaid continents. In Africa and Oceania, PCV2a was indeed the most prevalent, although the amount of data available were scant and, hence, the results were not representative.
Despite the evolution of PCV2, it is considered that the immune response, both humoral and cellular, has the capacity to give rise to a cross-reaction among the different genotypes (Semadaali et al. 2015, Fort et al. 2012).
On the other hand, it has not been demonstrated that PCV2 virulence may have varied during this evolution (Franzo et al. 2020), owing to which it cannot be concluded that there are some strains that are more virulent than others.
It is known that, in viruses in general, recombination may contribute to the evasion of the host immune response (Pérez-Losada, 2015). Nonetheless, there is no evidence of the existence of recombinant strains of PCV2 which have successfully escaped the protection offered by current vaccination programmes. Moreover, it should not be overlooked that there is currently no consensus in the scientific community on the existence of recombinant strains. Thus, we can conclude that it is currently not known whether recombination, as a mechanism for the evolution of the virus, may jeopardize the current vaccination plans.
Based on the current knowledge of the virus , experts on PCV2 sustain that recombination is a source of genetic variation not sufficiently strong to affect the protection capacity of current vaccines (Segalés, 2020).
A vaccine is much more than an antigen placed in a vial. Several other aspects, such as adjuvant and administration route, are fundamental when defining the immune response that the vaccine is going to produce.
Additionally, the formulation of the vaccine is also conditioned by safety aspects and other characteristics and requirements demanded by customers and authorities. For example, safety issues, protection against different genotypes of PCV2 or combination with vaccines for other diseases, as in the case of combinations with Mycoplasma hyopneumoniae.
Even though vaccines may produce a crossed immune response to different genotypes of PCV2, this does not guarantee that they can offer protection
Indeed, to date, it has not been possible to identify any marker that can accurately demonstrate whether an immunised animal is protected or not. Consequently, clinical efficacy trials, under either field or experimental conditions, are absolutely necessary to demonstrate protection against the different genotypes and thus meet the requirements of the authorities for inclusion into their indications. Of all the vaccines registered for PCV2, only a small number have shown their clinical efficacy against different genotypes of PCV2, and thus have had this indication registered.
Therefore, any PCV2 vaccine might have the potential to prevent disease in pigs infected with certain strains of PCV2d; however, unless indicated, this does not mean that it can control infection by PCV2d in all situations, and even less so in situations with a medium to high level of challenge.
Mhyosphere PCV® ID
Mhyosphere® PCV ID is the first and only vaccine based on a recombinant strain of Mycoplasama hyopneumoniae, called Nexyhon, which expresses the PCV2 capsid protein in its cytoplasm.
Mhyosphere® PCV ID demonstrated, under field conditions, to be able offering the following benefits:
- Control of Enzootic pneumonia, with reduction of lung lesions due to Mycoplasma hyopneumoniae and their incidence on farm.
- Broad protection against PCV2 genotypes: Mhyosphere® PCV ID was the first vaccine in Europe with licensed efficacy against the most prevalent genotypes of PCV2 (a, b and d). Reduces viraemia, virus load in lungs and lymphoid tissues and the duration of the viraemic period.
- Reduction of the losses of average daily weight gain and mortality caused by the disease, as observed in field studies at an age of 6 months.
Take home messages:
- In the case of PCV2, a number of cases of recombination have been described, although the reliability thereof is questioned by the scientific community.
- Experts on PCV2 sustain that recombination is a source of genetic variation not sufficiently strong to affect the protection capacity of current vaccines.
- PCV2a, PCV2b and PCV2d are the most widespread genotypes, accounting for 96.31% of the sequenced PCV2 genomes in domestic pigs.
- Even though vaccines may produce a crossed immune response to different genotypes of PCV2, this does not guarantee that they can offer protection.
- Clinical efficacy trials, under either field or experimental conditions, are absolutely necessary to demonstrate protection against the different genotypes and thus meet the requirements of the authorities for inclusion into their indications
| References | ||||
|---|---|---|---|---|
| 1. Firth C, Charleston MA, Duffy S, Shapiro B, Holmes EC. Insights into the evolutionary history of an emerging livestock pathogen: porcine circovirus 2. J Virol. 2009 Dec;83(24):12813-21. doi: 10.1128/JVI.01719-09. Epub 2009 Oct 7. PMID: 19812157; PMCID: PMC2786836. | ||||
| 2. Fort M, Sibila M, Nofrarías M, Pérez-Martín E, Olvera A,Mateu E, Segalés J. Evaluation of cell-mediated immune responses against porcine circovirus type 2 (PCV2) Cap and Rep proteins after vaccination with a commercial PCV2 sub-unit vaccine. Vet Immunol Immunopathol. 2012 Nov 15;150(1-2):128-32. doi: 10.1016/j.vetimm.2012.09.001. Epub 2012 Sep 5. PMID: 23010221. | ||||
| 3. Franzo G, Segalés J. Porcine circovirus 2 (PCV-2) genotype update and proposal of a new genotyping methodology. PLoS One. 2018;13(12):e0208585. Published 2018 Dec 6. doi:10.1371/journal.pone.0208585 | ||||
| 4. Franzo G, Segalés J. Porcine Circovirus 2 Genotypes, Immunity and Vaccines: Multiple Genotypes but One Single Serotype. Pathogens. 2020 Dec 14;9(12):1049. doi: 10.3390/pathogens9121049. PMID: 33327478; PMCID: PMC7764931. | ||||
| 5. Ssemadaali MA, Ilha M, Ramamoorthy S. Genetic diversity of porcine circovirus type 2 and implications for detection and control. Res Vet Sci. 2015 Dec;103:179-86. doi: 10.1016/j.rvsc.2015.10.006. Epub 2015 Oct 23. PMID: 26679815. | ||||
| 6. Kwon, T.; Lee, D.; Yoo, S.; Je, S.; Shin, J. and Lyoo, Y. (2017). Virus Res. 228:24-29. | ||||
| 7. Pérez-Losada M, Arenas M, Galán JC, Palero F, GonzálezCandelas F. Recombination in viruses: mechanisms, methods of study, and evolutionary consequences. Infect Genet Evol. 2015;30:296-307. doi:10.1016/j. meegid.2014.12.022 | ||||
| 8. Segalés, 2020. https://www.3tres3.com/articulos/%C2%BFque-relevancia-tienen-las-diferenciasentre-secuencias-de-pcv-2_45012/ | ||||
| 9. Guo LJ, Lu YH, Wei YW, Huang LP, Liu CM. Porcine circovirus type 2 (PCV2): genetic variation and newly emerging genotypes in China. Virol J. 2010;7:273. Published 2010 Oct 19. doi:10.1186/1743-422X-7-273 | ||||
| 10. Dupont K, Nielsen EO, Baekbo P, Larsen LE. Genomic analysis of PCV2 isolates from Danish archives and a current PMWS case-control study supports a shift in genotypes with time. Vet Microbiol. 2008 Apr 1;128(1- 2):56-64. doi: 10.1016/j.vetmic.2007.09.016. Epub 2007 Oct 2. PMID: 17996404. | ||||
| 11. Segalés J. Porcine circovirus type 2 (PCV2) infections: clinical signs, pathology and laboratory diagnosis. Virus Res. 2012 Mar;164(1-2):10-9. doi: 10.1016/j.virusres.2011.10.007. Epub 2011 Oct 17. PMID: 22056845 | ||||







