



McKean Conf: Genetic resistance for F18 Escherichia coli holds
Researchers observe increase in cases of post-weaning diarrhea cause by E. coliEditor's note: The following is taken from the James D McKean Swine Disease Conference held in July 2024 in Ames, Iowa. It is from a presentation given by Dr. Lucina Galina with PIC and Rodrigo Paiva and Marcelo Almeida with Iowa State University's Veterinary Diagnostic Laboratory.
Escherichia coli is a type of gram-negative bacteria that can cause disease in pigs, such as neonatal and post-weaning diarrhea, septicemia, polyserositis, arthritis, mastitis, urogenital infections, and edema disease. The Iowa State University Veterinary Diagnostic Laboratory (ISU-VDL) has observed an increase in cases of post-weaning diarrhea caused by E. coli from 2019 through 2023.
Genotyping conducted on a subset of post-weaning colibacillosis cases from previous years (2010-2018) indicated a balance between F18 and K88 fimbrial types, with 56.9% (range: 48.8% to 63.8%) of isolates positive for F18 and 43.1% (range: 36.2% to 51.2%) for K88. However, during the period of increased post-weaning colibacillosis diagnoses, F18 E. coli has become the predominant fimbrial type in 77.9% (range: 63.8% to 88.8%) of cases. Various reasons have been speculated for the heightened activity of F18 E. coli, including changes in antimicrobial practices due to market restrictions and stricter regulations, increased antimicrobial resistance, higher virulence in contemporary isolates, and the failure of immunological interventions (avirulent vaccines or competitive exclusion products) to prevent disease expression.
Another hypothesis was the genetic susceptibility of specific breeds to F18 E. coli. The expression of specific receptors on enterocytes for the attachment of F18 E. coli is regulated by the FUT1 gene. A point mutation G>A at base-pair 307 in the FUT1 gene can influence the expression of the receptor for F18 fimbrial attachment. Pigs with the FUT1AA genotype are resistant, while pigs with the FUT1GA or FUTGG genotype are susceptible to colonization by F18 E. coli. Therefore, this study aimed to investigate the effect of challenge with a contemporary F18 E. coli isolate of high virulence in pigs of resistant and susceptible genotypes from two different genetic pig lines.
Materials and methods
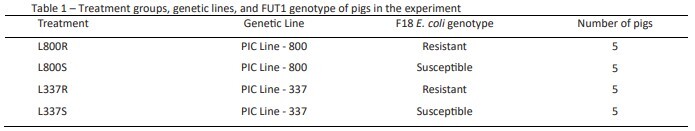
Twenty 21-day-old pigs were sourced from PIC genetics with specific FUT1 genotypes and genetic lines, as shown in Table 1. PIC performed FUT1 genotyping to select pigs before shipment to ISU. At arrival, pigs were placed in two different rooms according to the genetic line and were weighed and acclimated for three days before the challenge.
Pigs with resistant and susceptible genotypes were housed in the same pens. After three days of acclimation (0 days post-infection, [dpi]), all pigs were inoculated with 10 mL (1.08 x 1010 CFU/ml) of an F18:LT:STa:STb:Stx2e wild-type isolates by oral gavage.
Pigs were weighed individually on dpi -3, 0, and 7. Rectal swabs were collected on dpi 0, 1, 2, 3, 5, and 7. The severity of diarrhea was evaluated daily with fecal score visually assessed using the following scale: 0 = solid, 1 = semi-solid, 2 = semiliquid, and 3 = liquid. A fecal score ≥ 2 was considered diarrhea. On dpi 7, all pigs were euthanized, and sections of the duodenum, proximal, mid, and distal jejunum, and ileum were collected and fixed in 10% neutral buffered formalin and refrigerated for histopathological evaluation and bacteriology culture.
The fecal swabs were plated onto TSA agar with 5% sheep blood and incubated overnight at 37°C. The hemolytic E. coli shedding was assessed based on a semiquantitative method measured using a 5-point scale ranging from 0 to 4 according to the number of streaked sections that had viable E. coli, where 0 corresponded to no growth, 1 corresponded to growth in the primary streak, 2 corresponded to growth extending into the secondary streak, 3 corresponded to growth into the tertiary streak, and 4 corresponded to growth into the quaternary section of the agar plate. The presence of hemolytic E. coli on individual colonies isolated was confirmed using Matrix-Assisted Laser Desorption/Ionization time-of-flight mass spectrometry (MALDI-TOF MS) technique.
One section of the duodenum, proximal, mid, and distal jejunum, ileum, and colon were evaluated histologically. Sections were evaluated for the presence of bacterial attachment consistent with E. coli morphology.
Results
All groups of pigs showed similar E. coli fecal shedding scores on days 0, 1, 2, 3, and 7. However, on day 5, the L337S group had a significantly higher fecal score than the L800R group, although it was not significantly different from the L800S and L337R groups. The average daily diarrhea score was statistically higher for pigs in the susceptible groups on days 2 (L800S only), 3 (L800S and L337S), 4 (L800S and L337S), 5 (L800S only), 6 (L800S only), and 7 (L800S and L337S) compared to the resistant pigs.
Although no statistical difference was observed between groups during the study period, a mortality rate of 40% (2 of 5) and 80% (4 of 5) was observed in groups L800S and L337S, respectively, while no mortality was observed for pigs in groups L800R and L337R. The weight at day 7 adjusted by the weight at day 0 was statistically higher for pigs in L800R (19.27 lbs) and L337R (19.02 lbs) than L800S (13.58 lbs) and L337S (15.14 lbs). In the assessment of E. coli colonization of the small intestines, a statistical difference was also noted.
Notably, none of the pigs in the resistant groups exhibited E. coli colonization in any of the microscopically assessed sections of the small intestine, while 90% of the pigs in the susceptible groups showed colonization in at least one section of the small
intestine.
Conclusion
In this study, pigs with resistant genotypes to F18 E. coli showed no signs of E. coli-related illness. In contrast, pigs with susceptible genotypes displayed clinical disease, mortality, and histological lesions of post-weaning colibacillosis, regardless of their genetic line.








